Chromatin | Inteins | Quorum Sensing| Photo-proxmity Labeling
Overview
We are interested in studying protein function by integrating the tools of synthetic organic and physical chemistry with those of molecular genetics. Driven by a series of biological questions, we have developed general chemical biology approaches that allow the covalent structure of proteins to be manipulated with a similar level of control to that possible with smaller organic molecules. These technologies, which can be applied both in vitro and in vivo, allow the insertion of unnatural amino acids, posttranslational modifications and isotopic probes site-specifically anywhere into proteins. Our methods are now used by numerous laboratories worldwide, and have allowed a large number of biomedical questions to be addressed. A summary of ongoing work in the Muir group is provided below:
Chromatin
The genomic DNA of eukaryotes is physically associated with an astonishing collection of nuclear factors, which serves to both store the nucleic acid in a stable form, chromatin, but also grant access to the information it encodes when needed. Understanding the mechanisms by which genomic access is controlled at the level of chromatin is thus key to unlocking how cellular phenotypes are established and maintained. Mistakes made in these epigenetic processes lead to the mis-expression (or mis-silencing) of genes with far-reaching implications for human biology and human disease such as cancer. Such epigenetic regulation is disarmingly complex and is known to involve the coordinated action of many players, including general and specific transcription factors, chromatin remodeling enzymes, non-coding RNAs and chemical modifications to both the DNA and the histone packaging proteins. One of great challenges in the field of epigenetics is to move beyond correlative type information, which is now in abundant supply, to the point where we can truly connect the dots at the molecular level. Establishing such causal relationships requires precise manipulation of the covalent structure of chromatin, a formidable undertaking. While chromosomes certainly present one of most challenging of all substrates for biochemical study, tremendous progress has been made during the last several years in the development of precision chemical biology tools for manipulating the covalent structure of chromatin, both in the test tube and in cells. Many of these approaches were developed in our group and have been used to define the molecular mechanisms underlying a number of fundamental epigenetic processes. For an overview of previous work in this area, see:
Hananya, N.; Koren, S.; Muir, T. W. Nat Rev Genet 2023. doi: https://doi.org/10.1038/s41576-023-00664-z
Interrogating epigenetic mechanisms with chemically customized chromatin.
Mueller, M.M. and Muir, T.W. 2015, Chemical Reviews, 115, 2296-2349
Histones: At the Crossroads of Peptide and Protein Chemistry
Ongoing work in our lab focuses on understanding how mutations to histone proteins themselves leads to various cancers, as well as how native chromatin structure is regulated intrinsically by post-translational modifications, and by the action of large molecular machines that remodel chromatin in the nucleus. To tackle these questions we are deploying several new approaches. These include a high-throughput biochemical platform that allows the massively parallel analysis of ‘designer’ chromatin libraries through the integration of advanced protein chemistry methods with those of next generation DNA sequencing. We are also developing protein-engineering methods for manipulating the chemical structure of cellular proteins and that allow native chromatin to be directly chemically customized. These new tools, which are designed to be complementary, provide a foundation for studying genomic regulation at the molecular level all the way from the test tube to the cell.
Inteins

Inteins are consummate molecular escape artists that spontaneously excise themselves, in a traceless manner, from proteins in which they are embedded. The posttranslational process these ‘Houdini’ proteins support is termed protein splicing. Inteins are extremely common proteins, being present in unicellular organisms from all domains of life, including several pathogens such as Mycobacterium tuberculosis (Mtb). Typically, inteins are embedded within essential proteins involved in DNA replication or repair, and thus protein splicing is required for cellular replication. While most inteins are contiguous polypeptides, a sub-set of these proteins are naturally fractured. These split inteins must first associate before protein trans-splicing can occur. Inteins, by virtue of the remarkably biochemical process they support, i.e. protein splicing, offer unique opportunities for discovering new mechanisms in biomolecular recognition and protein-based catalysis. Indeed, the study of protein splicing has led to the development of a suite of widely used chemical biology technologies for engineering proteins. These intein-based technologies effectively bridge the worlds of chemistry and biology, providing investigators with a means to traverse the disciplines by applying chemistry to proteins. For an overview of this area, see:
Thompson, R.E.; Muir, T.W. Chem Rev. 2020. 120(6), 3051-3126. doi: 10.1021/acs.chemrev.9b00450 Chemoenzymatic Semisynthesis of Proteins
Debelouchina, G.T. and Muir, T.W. 2017, Quarterly Reviews in Biophysics, 50, E7. A molecular engineering toolbox for the structural biologist
Ongoing work in our lab focuses on the study of the naturally split inteins. Recently described members of this family exhibit remarkably fast protein splicing rates (seconds). Moreover, the intein fragment associate tightly (low nanomolar affinities) and with high specificity. At a fundamental level, we aim to characterize how these ‘ultrafast’ split inteins recognize one another and catalyze a multi-step reaction cascade so quickly. This information will aid in the discovery of additional ‘ultrafast’ inteins from meta-genomic analyses, as well as in the development of new technologies. With respect to the latter, we currently have significant interest in developing methods that allow split intein activity to be controlled and tuned in cells, thereby feeding our interests in chromatin with powerful new cell-based tools.
Quorum Sensing
The ability of bacteria to coordinate group activity at particular phases of growth is essential for many fundamental processes such as competence, bioluminescence, biofilm formation, and virulence. This feat is achieved by quorum sensing (QS) signaling mechanisms that allow an individual bacterium to sense the density of the entire population of which it is a member. We have defined the molecular structure of a family of secreted peptides from S. aureus that control virulence in the organism through a conserved QS signaling pathway termed agr. Agr remains the best-characterized quorum sensing pathway in any Gram-positive organism and, given its biomedical importance, is now widely studied. Using a combination of c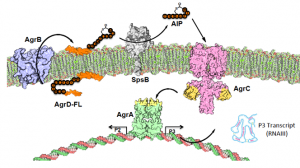 hemistry, protein engineering and molecular genetics, we have figured out many aspects of the molecular mechanism of this critical process. This understanding has, among other things, led to the rational design of global inhibitors of virulence in S. aureus that prevent infections in animal models and that thus have therapeutic potential. For a recent overview of this area, see:
hemistry, protein engineering and molecular genetics, we have figured out many aspects of the molecular mechanism of this critical process. This understanding has, among other things, led to the rational design of global inhibitors of virulence in S. aureus that prevent infections in animal models and that thus have therapeutic potential. For a recent overview of this area, see:
Wang, B. and Muir, T.W. 2016, Cell Chemical Biology, 23, 214-224 Regulation of Virulence in Staphylococcus aureus: Molecular Mechanisms and Remaining Puzzles.
Our current work builds on recent breakthroughs that have allowed us to reconstitute much as the quorum sensing circuit using purified components. We are particularly focussed on obtaining a deeper understanding into the molecular processes underlying the production and sensing of the autoinducer peptide (AIP) pheromone that is central to agr regulation. These studies combine small molecule synthesis methods with those of structural biology, chemical biology and microbial genetics. These studies will fuel the development of new therapeutic strategies targeting agr, but also contribute to a fundamental understanding of QS systems of this type, which are pervasive in gram-positive bacteria.
Photo-proximity Labeling
Hananya, N.*, Ye, X.*, Koren, S.; Muir, T. W. A genetically encoded photoproximity labeling approach for mapping protein territories. Proceedings of the National Academy of Sciences 2023 120(16) e2219339120
Seath, C.P.*, Burton, A.J.*, Sun, X.; Lee, G.; Kleiner, R. E.; MacMillan, D. W. C.; Muir, T. W. Tracking chromatin state changes using nanoscale photo-proximity labelling. Nature 2023 616 574-580